Elemental Hub's WP4 lead, Joanne Santini and member of our Strategic Advisory Board, Karen Hudson-Edwards collaborated with the following scholars on a publication that is relevant to WP1: Katherine R. Lane, Sarah E. Jones, Thomas H. Osborne, David Geller-McGrath, Bennet C. Nwaobi, LinXing Chen, Brian C. Thomas, and Jillian F. Banfield.
ABSTRACT
Copper bioleaching is a green technology for the recovery of copper from chalcopyrite (CuFeS2) and chalcocite (Cu2S) ores. Much remains to be learned about how mineral type and surface chemistry influence microbial community composition. Here, we established a microbial consortium from a copper bioleaching column in Cyprus on chalcopyrite and then sub-cultured it to chalcocite to investigate how the community composition shifts due to changes in mineral structure and the absence of mineral-derived Fe. The solution chemistry was determined and microbial communities characterised by genome-resolved metagenomics after 4 and 8 weeks of cultivation. Acidithiobacillus species and strains, a Rhodospirilales, Leptospirillum ferrodiazotrophum and Thermoplasmatales archaea dominated all enrichments, and trends in abundance patterns were observed with mineralogy and surface-attached versus planktonic conditions. Many bacteria had associated plasmids, some of which encoded metal resistance pathways, sulphur metabolic capacities and CRISPR-Cas loci. CRISPR spacers on an Acidithiobacillus plasmid targeted plasmid-borne conjugal transfer genes found in the same genus, likely belonging to another plasmid, evidence of intra-plasmid competition. We conclude that the structure and composition of metal sulphide minerals select for distinct consortia and associated mobile elements, some of which have the potential to impact microbial activity during sulphide ore dissolution.
Graphical Abstract
Bioleaching is a microbially mediated green technology for the recovery of metals from ore. Here, microbial consortia from a copper bioleaching column in Cyprus were cultivated on chalcopyrite (CuFeS2) and then sub-cultured on chalcocite (Cu2S) and chalcopyrite to investigate how mineral type and surface chemistry influence microbial community composition.
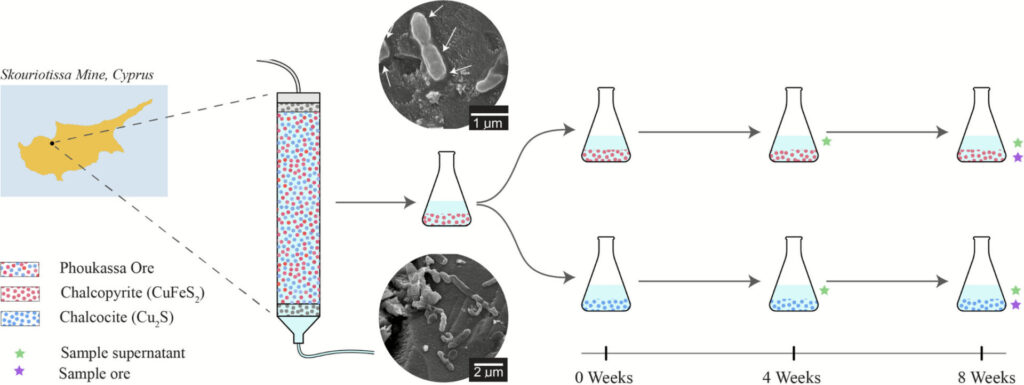
Schematic of metagenomic experimental design. The diagram illustrates the experimental design of the project. (a) Microbial consortium was sampled from the packed bioleaching column ‘SC3’ containing Phoukassa ore which has a natural mixture of chalcopyrite (red) and chalcocite (blue) at the Skouriotissa Mine in Cyprus. (b) The microbial community was cultivated and sub-cultured in minimal acid medium with chalcopyrite 10 times. (c) Microbial community was sub-cultured in minimal acid medium with chalcopyrite or chalcocite, and subsequently sub-cultured on the same mineral five times prior to samples taken for DNA isolations. Metagenomes were sequenced from the supernatant at 4 weeks (green star), and from the supernatant (green star) and ore (blue star) separately at 8 weeks, totalling six metagenomes. Each metagenome consisted of DNA pooled from three replicates. Abiotic controls for each treatment consisted of minimal acid medium containing chalcopyrite or chalcocite (without the microbial inoculum) were sampled for chemical analysis at weeks 0, 4 and 6 to measure background abiotic dissolution processes.